The Brain Imaging Data Structure (BIDS) project is an effort to create data standards for accessibility, usability and reproducibility of neuroimaging data. Initially developed for MRI data it has now been extended to EEG as BIDS-EEG.
As neuroscience makes an effort to solve issues in reproducibility one of the key challenges is the lack of standards in the way data is captured, processed and stored and the consequent difficulty in the ability to both reproduce findings as well as enable more easy sharing of data between labs. BIDS evolved in the MRI community as an effort to solve these issues. Fundamentally what it is involves a set of standards for how to structure and name data files and ensure that all key data elements are included.
The Basics of BIDS for EEG
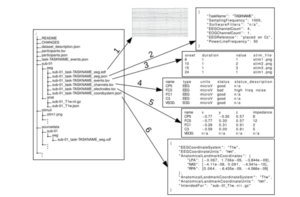
BIDS specifies various aspects from the way in which EEG files should be stored (there are .edf is the preferred format, followed by to how the data should be organized. Other than the EEG file itself, BIDS specifies the file structure and metadata to be provided for each subject. For each subject for instance there would be a directory with the following elements:
- EEG raw data for each session (European Data format or .edf and BrainVision Core Data Format are the official types though .edf is preferred)
- raw data from any other modality recorded for each session (e.g. EOG)
- File listing the channels and/or electrodes saved as “channels.tsv” or “electrodes.tsv” file
- Json file “coordsystem.json” providing the coordinate system file.
- Additional metadata file(s) which exhaustively specifies all the details of the experimental task and EEG recording system in json format (.json)
- A stimuli directory (if part of the experiment)
- A code directory to enable any required data conversion and preprocessing to be reproduced.
An example is provided below (from [1]
You can also watch a video about it by Dr.Cyril Pernet, one of the key BIDS-EEG architects here:
Some key clarifications
Channels vs Electrodes: BIDS-EEG distinguishes between electrodes and channels as follows: An electrode is a contact point attached to the skin while a channel is the combination of the analog differential amplifier and analog-to-digital converter that result in a potential (voltage) difference being stored in the EEG dataset. The “reference” and “ground” electrodes should therefore not be referred to as channels and only as electrodes.
Fiducial vs Anatomical Landmarks: BIDS-EEG defines fiducials as objects with a well defined location used to facilitate the localization of electrodes and co-registration with some kind of head image or model (e.g.Vitamin E pills which are visible in the MR image) while anatomical landmarks represent locations on a research subject such as the nasion (intersection of the frontal bone and two nasal bones of the human skull).
The challenges of BIDS for EEG
Neuroimaging methods such as MRI and fMRI utilize very large and expensive equipment and the number of manufacturers and models available are therefore fewer. Consequently there are fewer hardware standards and file. In contrast there are numerous EEG hardware manufacturers and models and an equally large number of configurations, internal preprocessing steps, reference choices and file formats. This makes the onus of organizing the data far more difficult as it often requires various scripts to convert from the manufacturers format into the BIDS format. The BIDS team is working to help researchers with this onerous task by building libraries of scripts to enable interoperability between platforms. There are projects well underway for reading BIDS formats within EEGlab and the MNE toolkit for example.
Ultimately however, despite the substantially more onerous task that converting data formats and organizing data structures presents for EEG relative to other neuroimaging methods, the effort is worthwhile. Without the effort of the EEG field to converge on standardized data formats and structures, the field will struggle to deliver on its potential.
BIDS Resources
[1] Pernet C et al EEG-BIDS, an extension to the brain imaging data structure for electroencephalography Nature Scientific Data June 2019
https://bids.neuroimaging.io – full BIDS-EEG specs
https://github.com/bids-standard/bids-starter-kit – community driven guides, tutorials and converter scripts
https://github.com/mne-tools/mne-bids – tools to read BIDS format in MNE


















