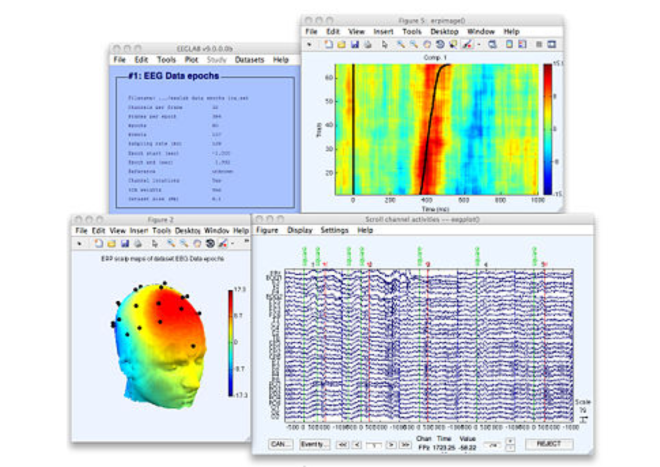
EEGLAB is an open source platform running on Matlab with a large set of functions for analyzing the EEG signal that are constantly being updatd. See what is new in the platform in this guest post from Arnaud Delorme.
EEGLAB is an interactive open source Matlab toolbox (The Mathworks, Inc.) for processing continuous and event-related EEG, MEG and other electrophysiological data that runs under Linux, Unix, Windows, and Mac OS X.
A background to EEGLAB
The EEGLAB signal processing environment began as a set of functions for electroencephalographic (EEG) data analysis and visualization released was released from the Swartz Center for Computational Neuroscience (SCCN) at the University of California San Diego (UCSD) in 2001. Currently, at least 102 EEGLAB plug-in tools or toolsets have been publicly released by researchers from many laboratories, and a worldwide survey of 687 cognitive neuroscientists by Hanke & Halcencko found EEGLAB to be the software environment most widely used for electrophysiological data analysis by a wide margin (neuro.debian.net/survey/2011/
Moving Towards Multimodal Brain Imaging Applications
One major shift in scientific perspective on the nature and use of human electroencephalographic (EEG) data is now ongoing. The application of and research into EEG algorithms and application of complex statistics to high-density EEG and to multimodal brain imaging data including EEG is currently limited by the relatively high computational demands posed by large and complex data sets. EEG analysis itself is also now rapidly moving toward becoming a three-dimensional source imaging modality by combining EEG analysis with models derived from detailed template and individual magnetic resonance and other structural head images. However, fully applying the power of new source-resolved human EEG and multimodal EEG data analysis and nonparametric statistical methods requires more computational power than many EEG laboratories have available. Full time/frequency analysis, transformation of high-density scalp EEG data to source-resolved independent component processes, more precise localization of these processes, and other new analysis steps such as general linear model estimation and associated statistics also are for many laboratories beyond the ‘compute horizon’ (i.e., requiring infeasibly long wait times). Our NIH supported effort has been to develop a platform to solve this problem.
Some new advances in EEGLAB
In this video, we show recent advance and support for new approaches to statistical analysis across subject groups, conditions, and task paradigms. The video below also shows progress in collaboration with the Neuroscience Gateway (nsgportal.org) project of the San Diego Supercomputer Center, of our Open EEGLAB Portal allowing users to run computationally intensive brain modeling and imaging processes on the academic high-performance computing (HPC) environment of the San Diego Supercomputer Center (sdsc.edu).
We believe that satisfactory progress toward these goals will keep the EEGLAB software environment relevant to the evolving research needs of its large user base, while also contributing to establishing source-resolved human brain electrophysiology as an important functional imaging method for research toward better understanding of relationships between distributed brain activity, behavior, and experience — in health, disease, and other physically or mentally challenging states and circumstances.